PyMDE: Minimum-Distortion Embedding¶
PyMDE is a Python library for computing vector embeddings of items, such as images, biological cells, nodes in a network, or any other type of abstract object. The embeddings are designed to minimally distort relationships between pairs of items, while possibly satisfying some constraints.
For more background about the methods that PyMDE uses, please see our monograph Minimum-Distortion Embedding, which introduces a simple but general framework for embedding.
You can use PyMDE to:
visualize datasets, small or large;
embed into arbitrarily large dimensions (much greater than 2 or 3);
generate feature vectors for supervised learning;
compress high-dimensional vector data;
draw graphs;
and more.
Unlike other popular embedding methods, PyMDE also lets you
compute large embeddings quickly, using a GPU;
fit embeddings that satisfy constraints, such as having uncorrelated feature columns;
design custom embeddings (and recreate well-known ones, like PCA, Laplacian eigenmap, LargeVis, and UMAP);
sanity-check embeddings using principled methods;
look for outliers in original data.
Example¶
Here’s an example that shows how to use PyMDE to visualize high-dimensional vectors (which in this case are images from the MNIST dataset).
import pymde
mnist = pymde.datasets.MNIST()
embedding = pymde.preserve_neighbors(mnist.data, embedding_dim=2, verbose=True).embed()
pymde.plot(embedding, color_by=mnist.attributes['digits'])

Each point represents an image in the dataset, and every point is colored by the digit represented by its underlying image. However, only the raw pixel data was used to create the embedding. This example embedded into two dimensions, allowing us to plot the result, but PyMDE supports embedding into much larger dimensions.
This is just one of the many things that PyMDE can do.
Why use PyMDE?¶
There are many reasons to try out PyMDE. Here are some.
PyMDE is expressive. PyMDE is based on a simple but general framework for embedding, called Minimum-Distortion Embedding (MDE). The MDE framework generalizes well-known methods like PCA, spectral embedding, multi-dimensional scaling, LargeVis, and UMAP. With PyMDE, it is easy to recreate well-known embeddings and to create new ones, tailored to your particular application.
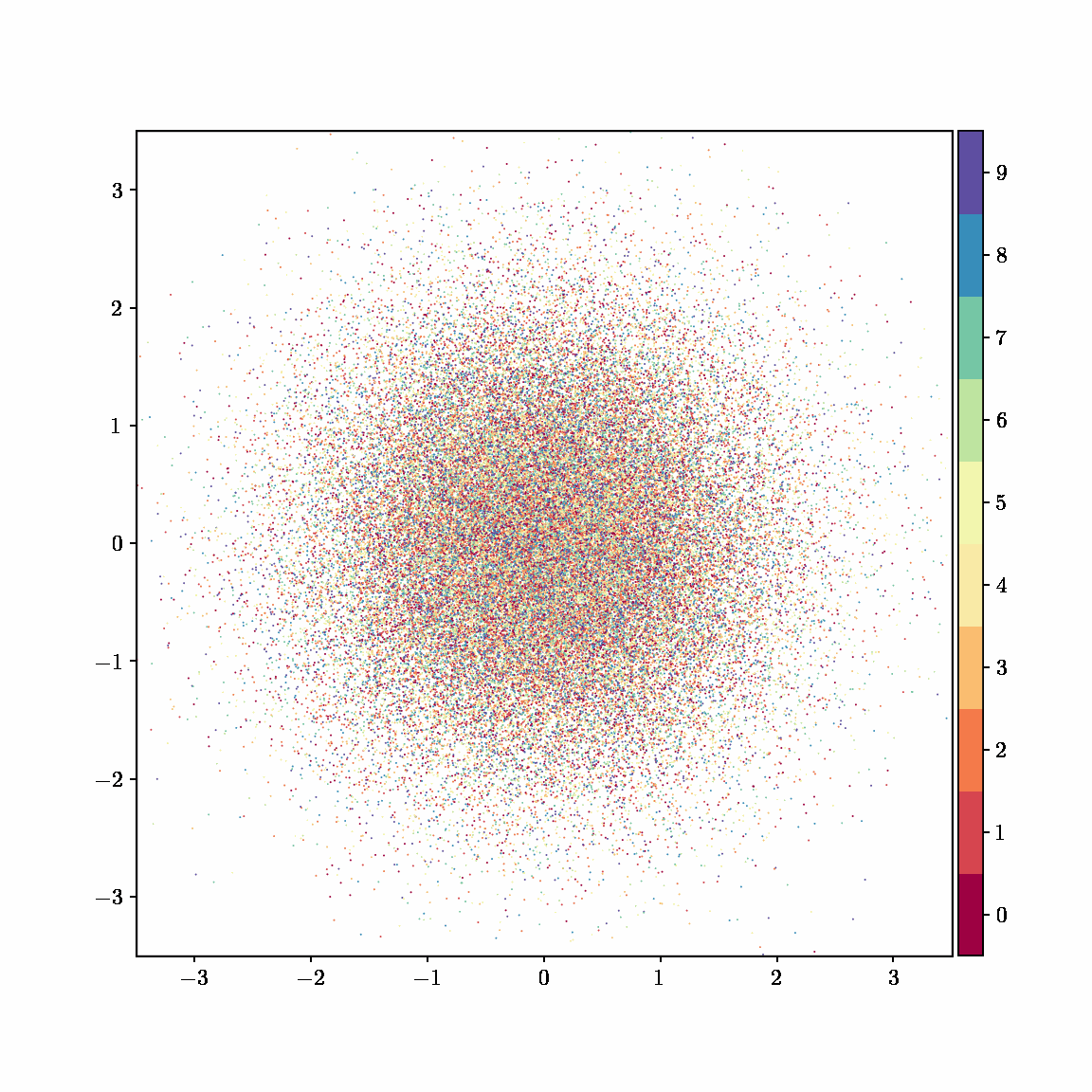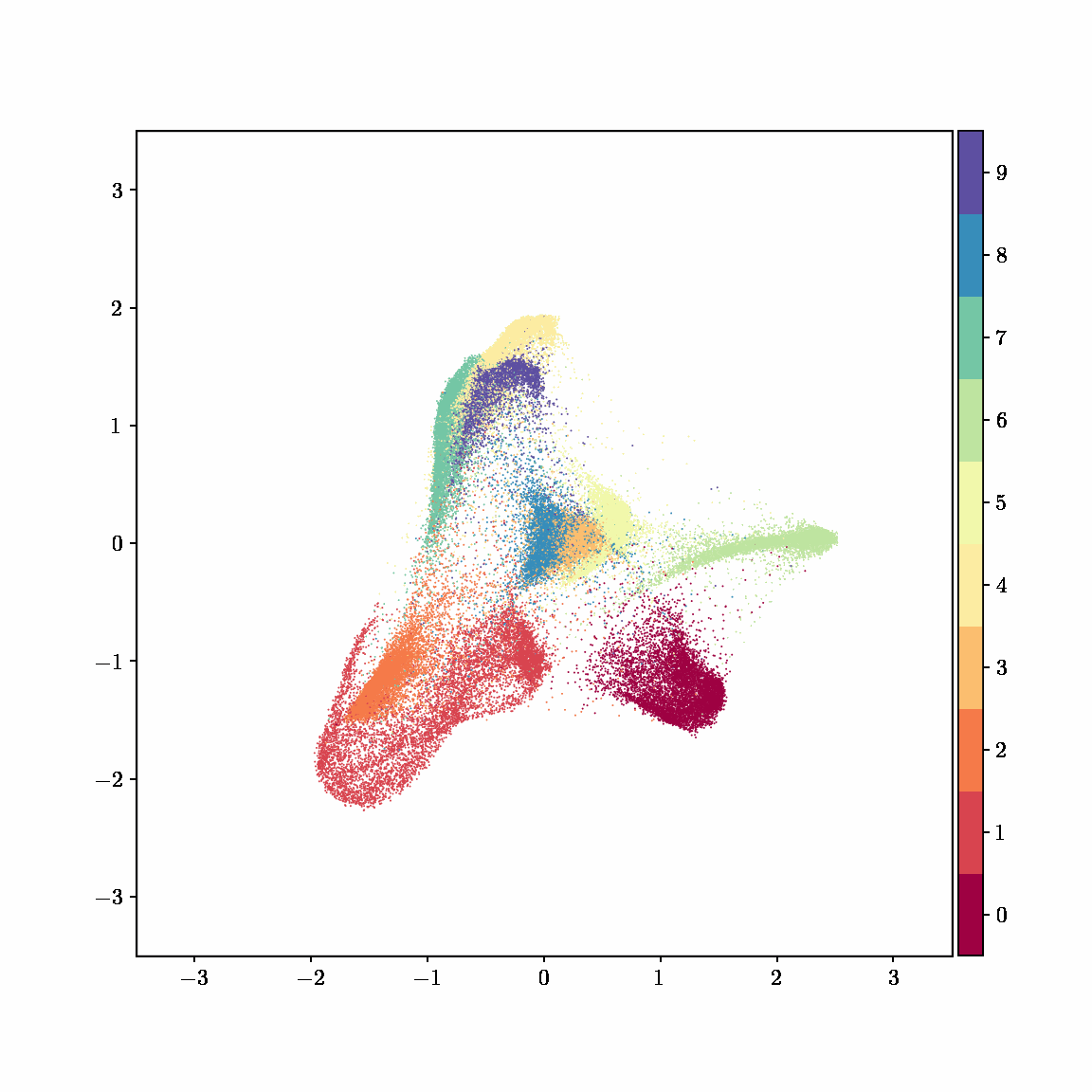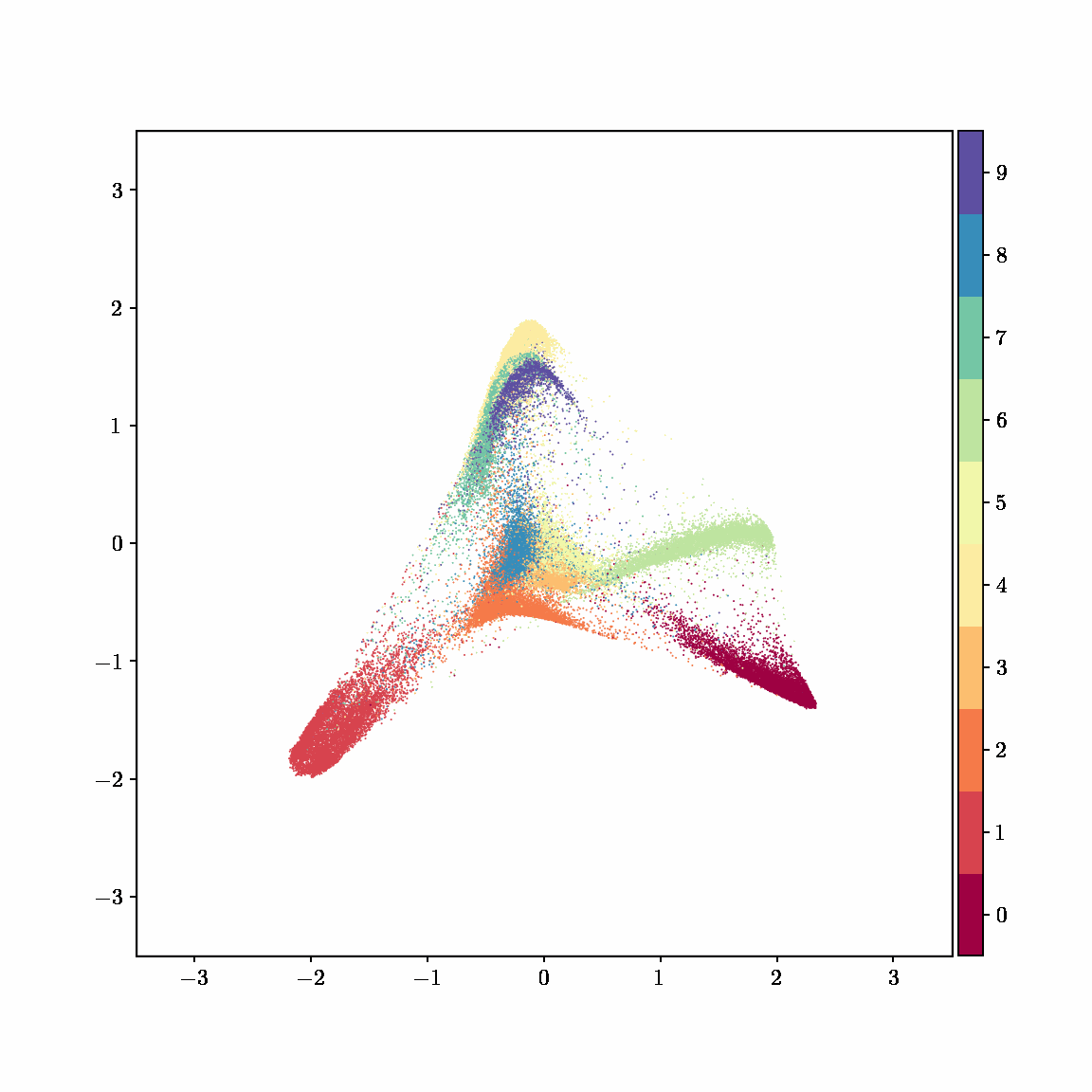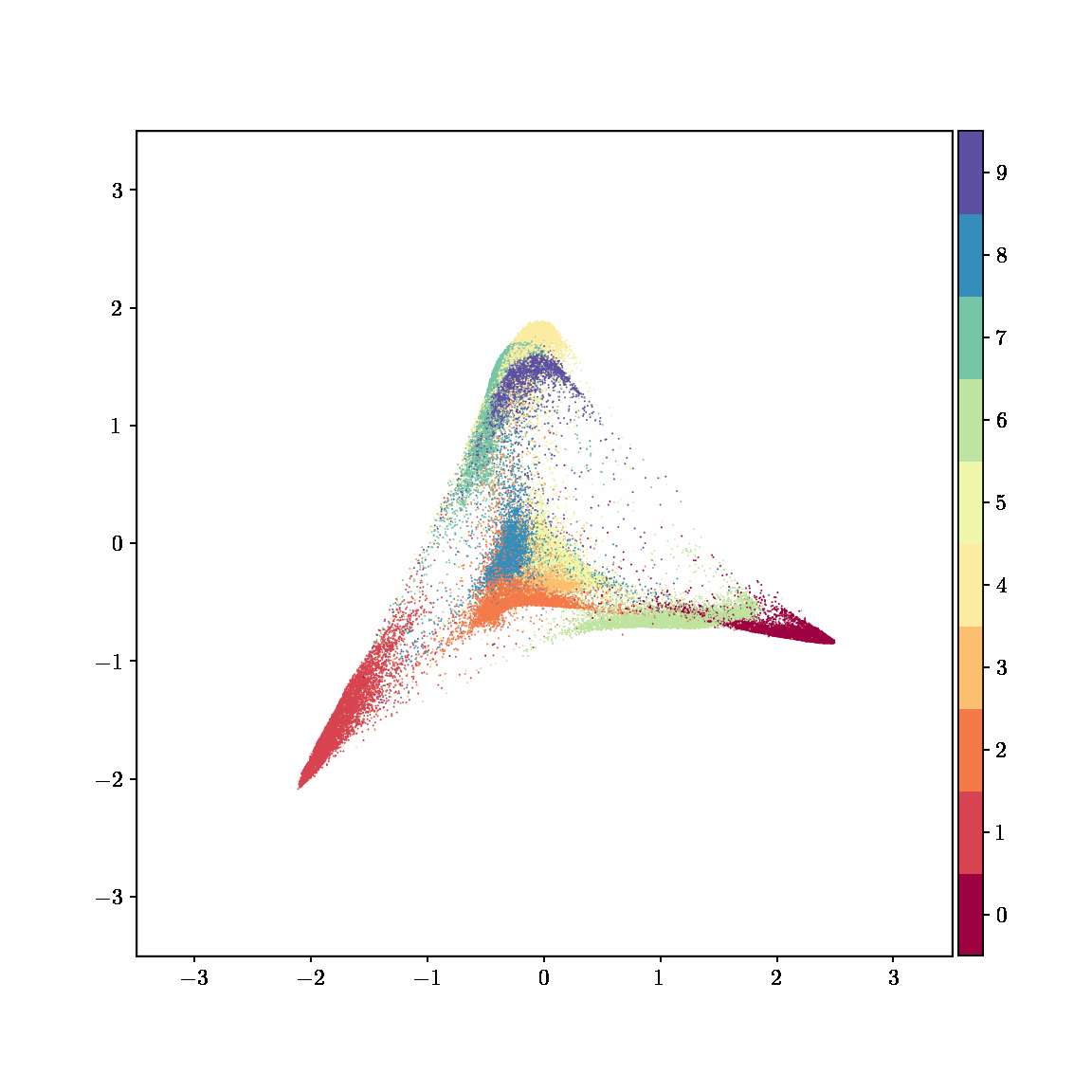
Unlike other embedding methods, PyMDE lets you impose constraints on the embedding. For example, the below GIFs show PyMDE computing three different kinds of embeddings of MNIST. The first and third embeddings are constrained to be standardized (which roughly means they have uncorrelated features), while the second is unconstrained. Standardized embeddings are favorably scaled for downstream machine learning tasks, such as supervised learning.
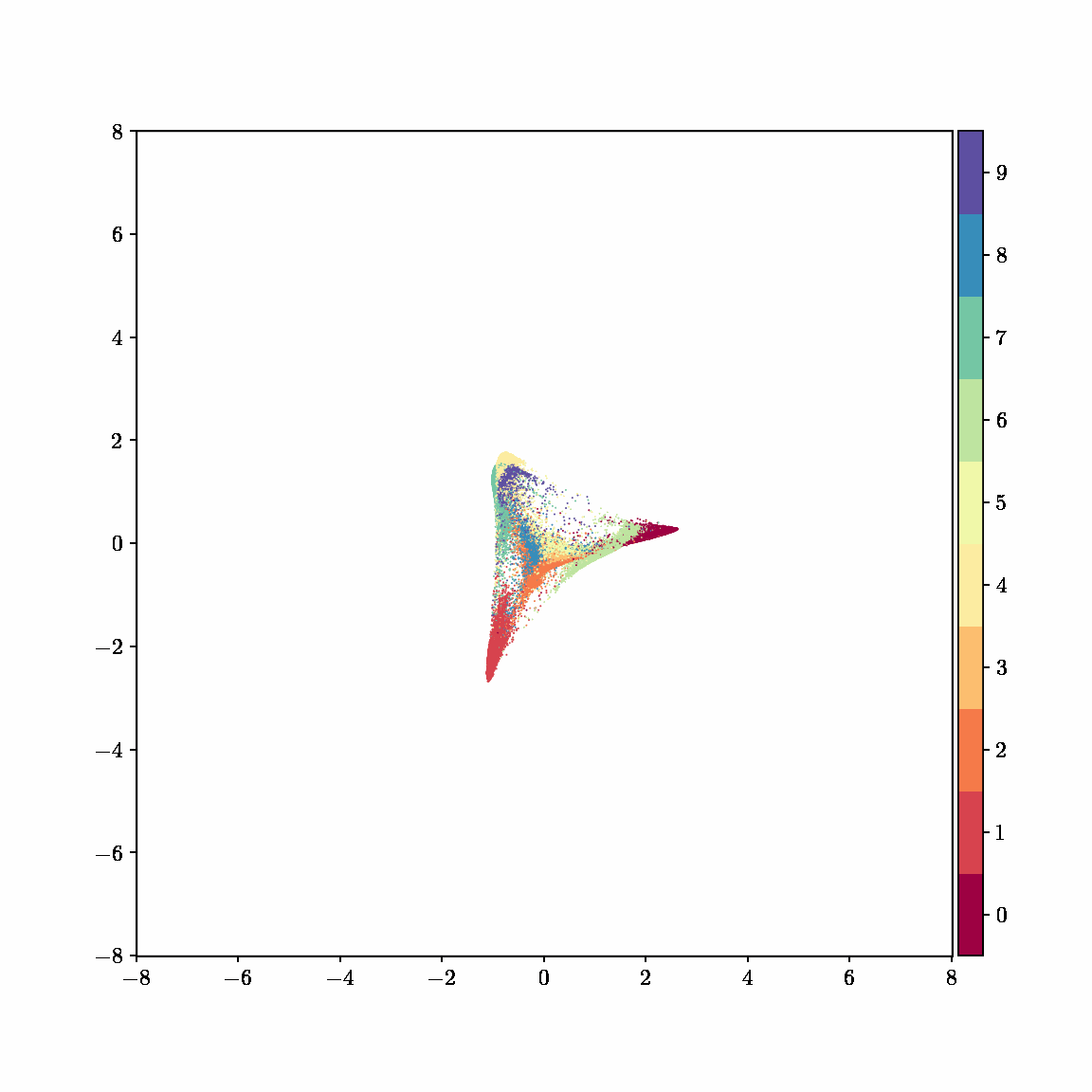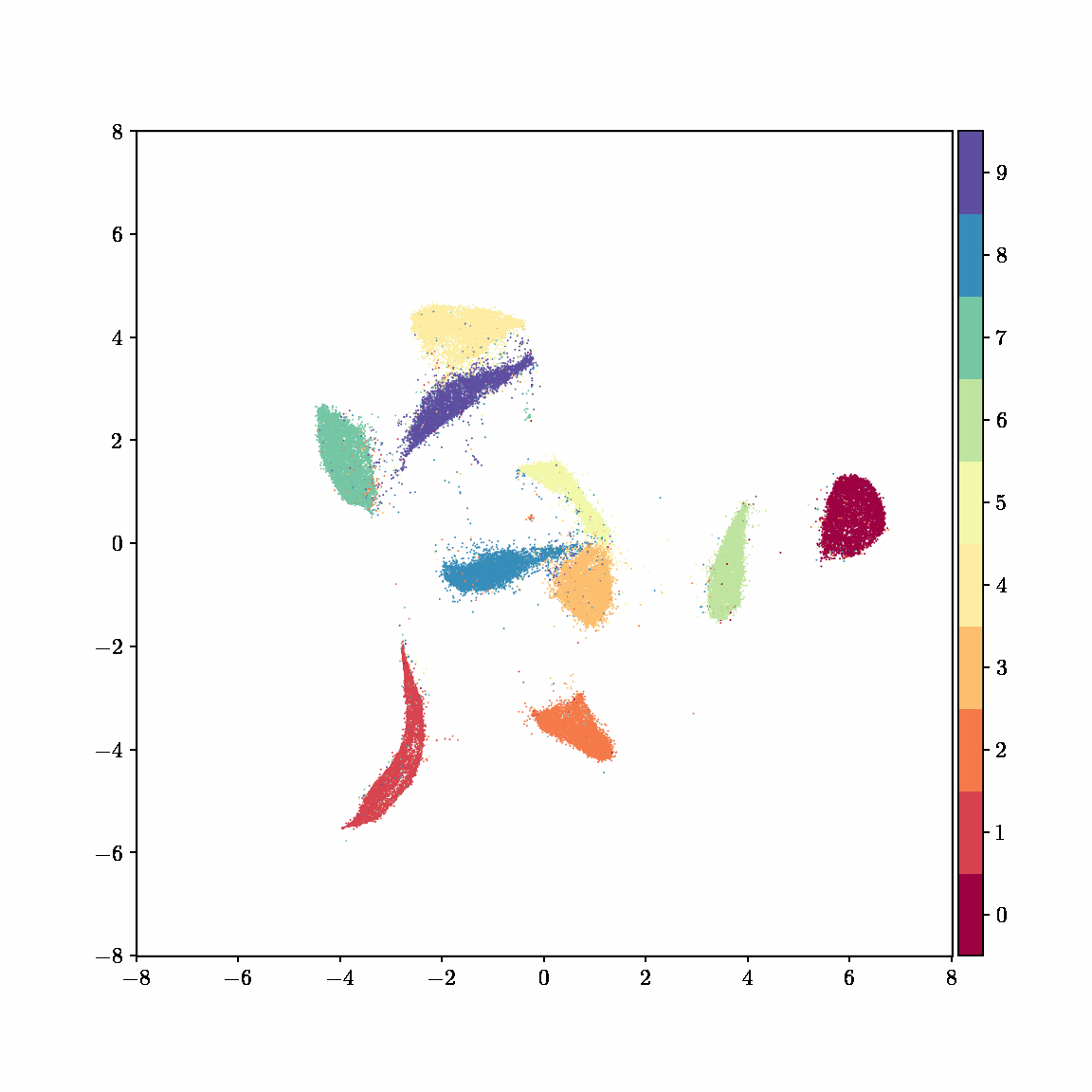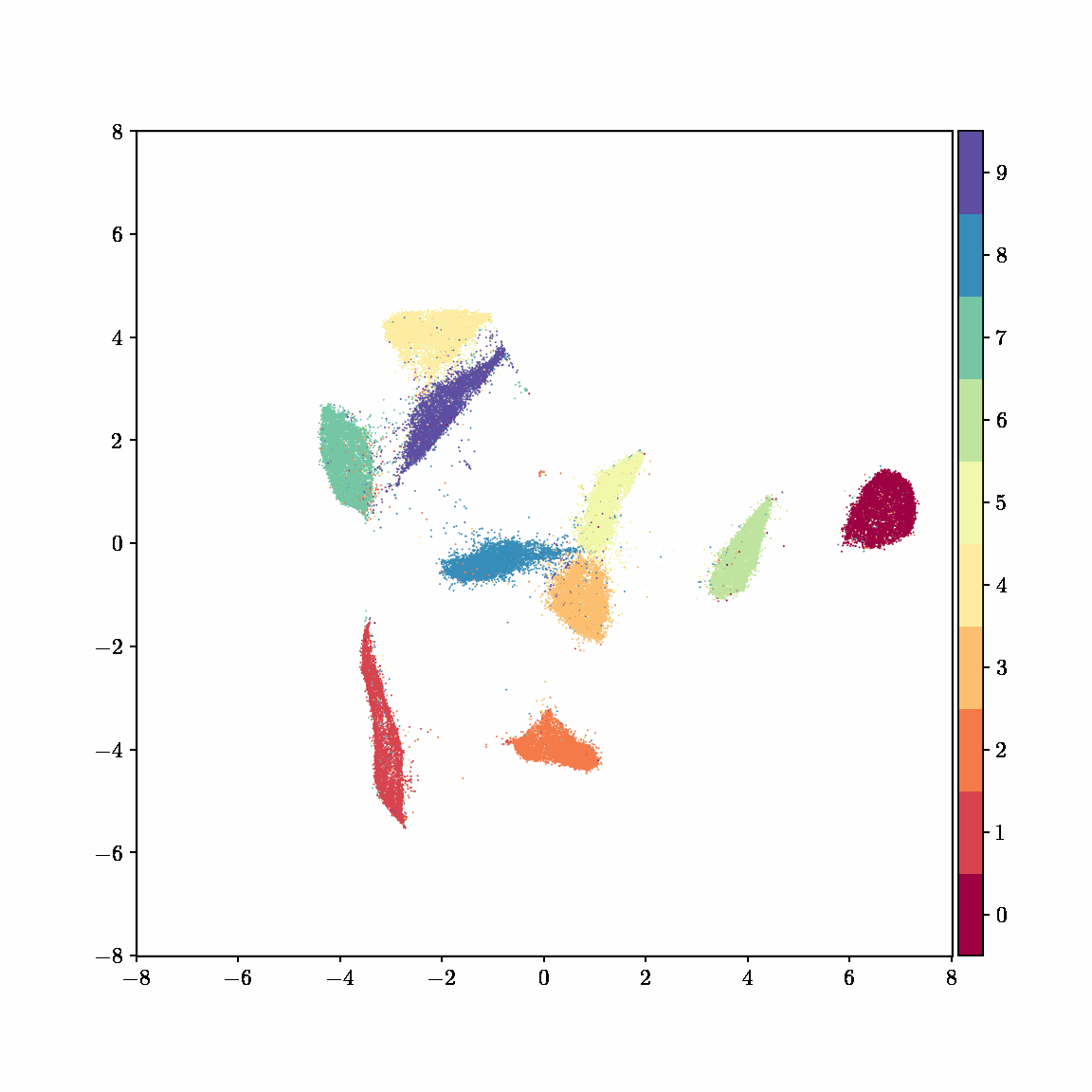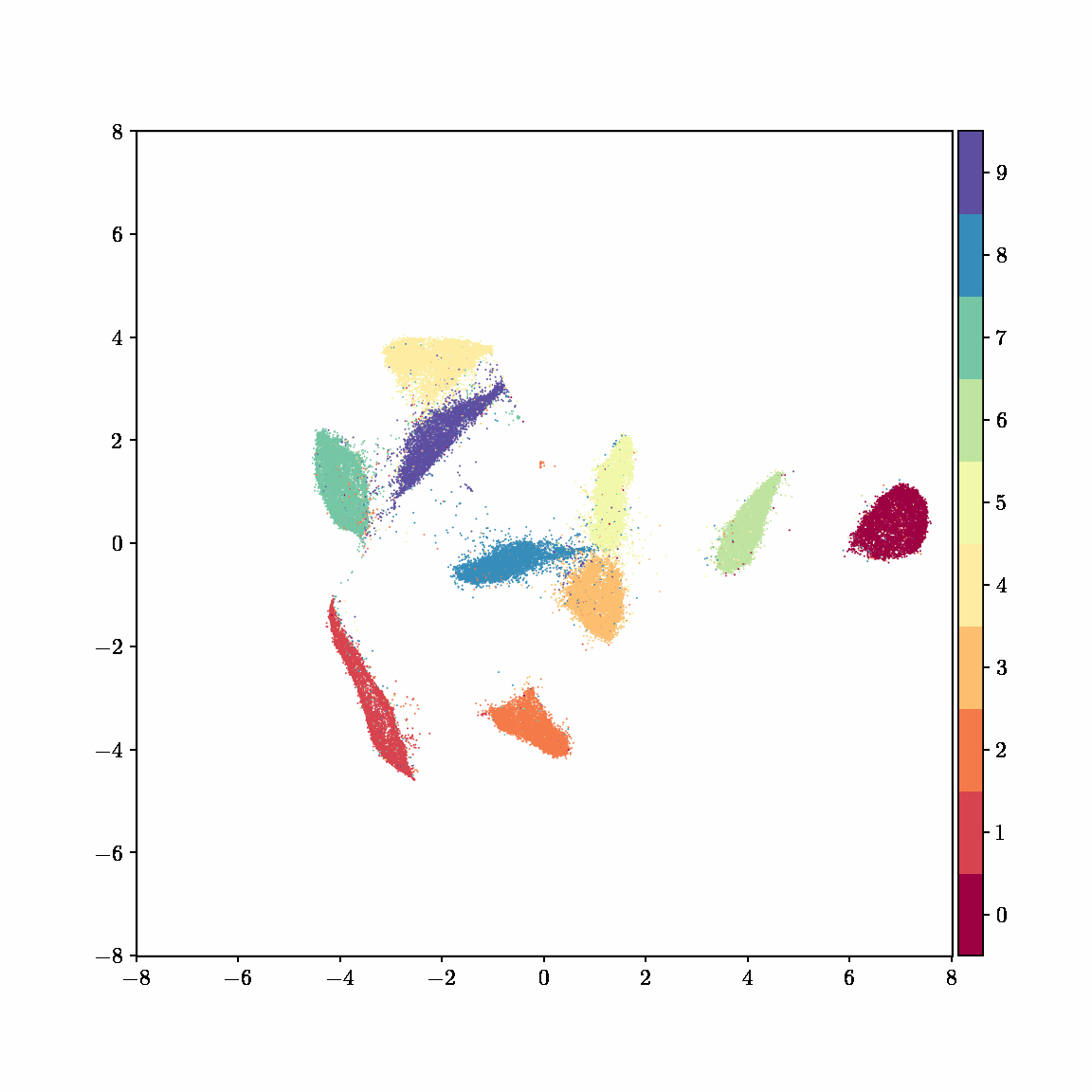
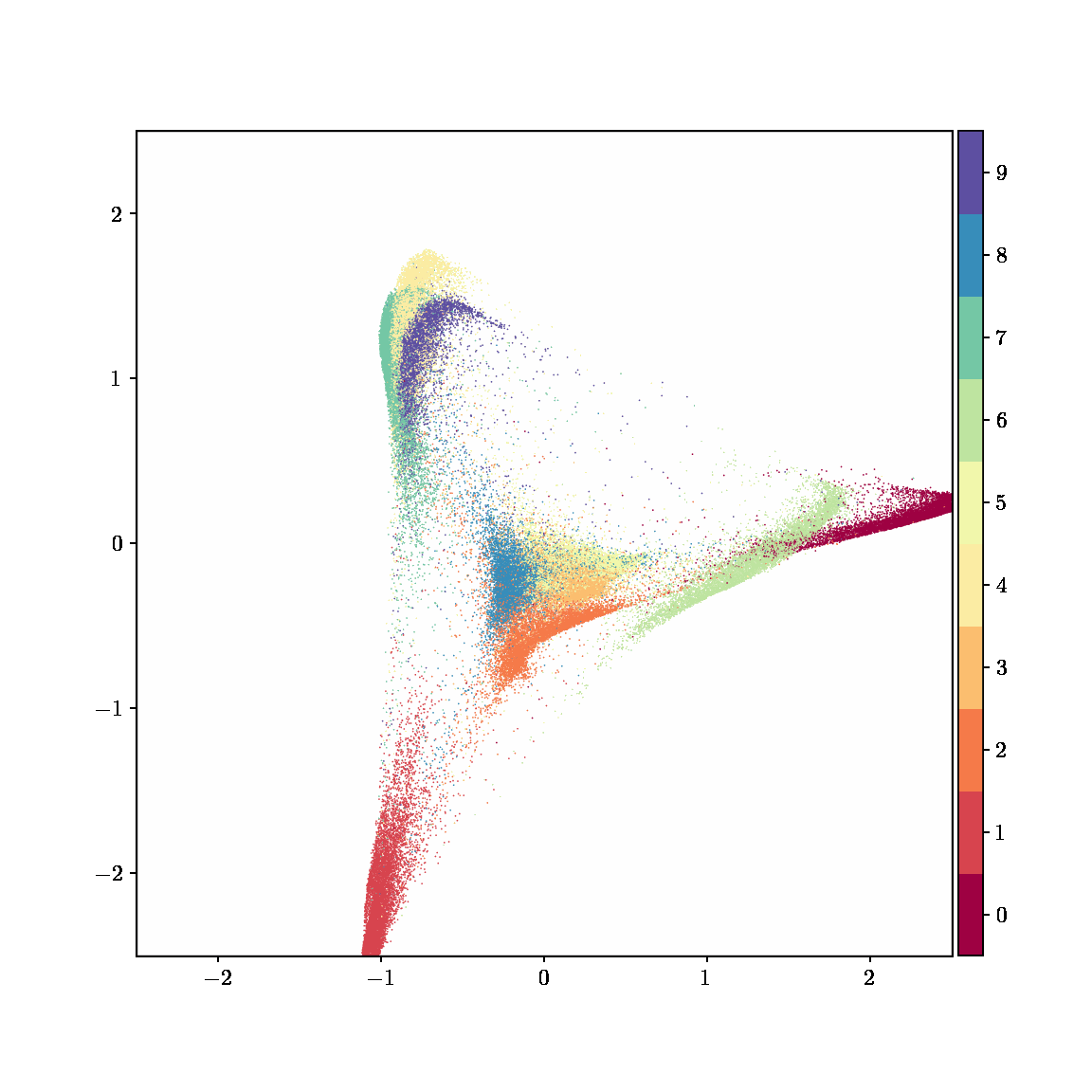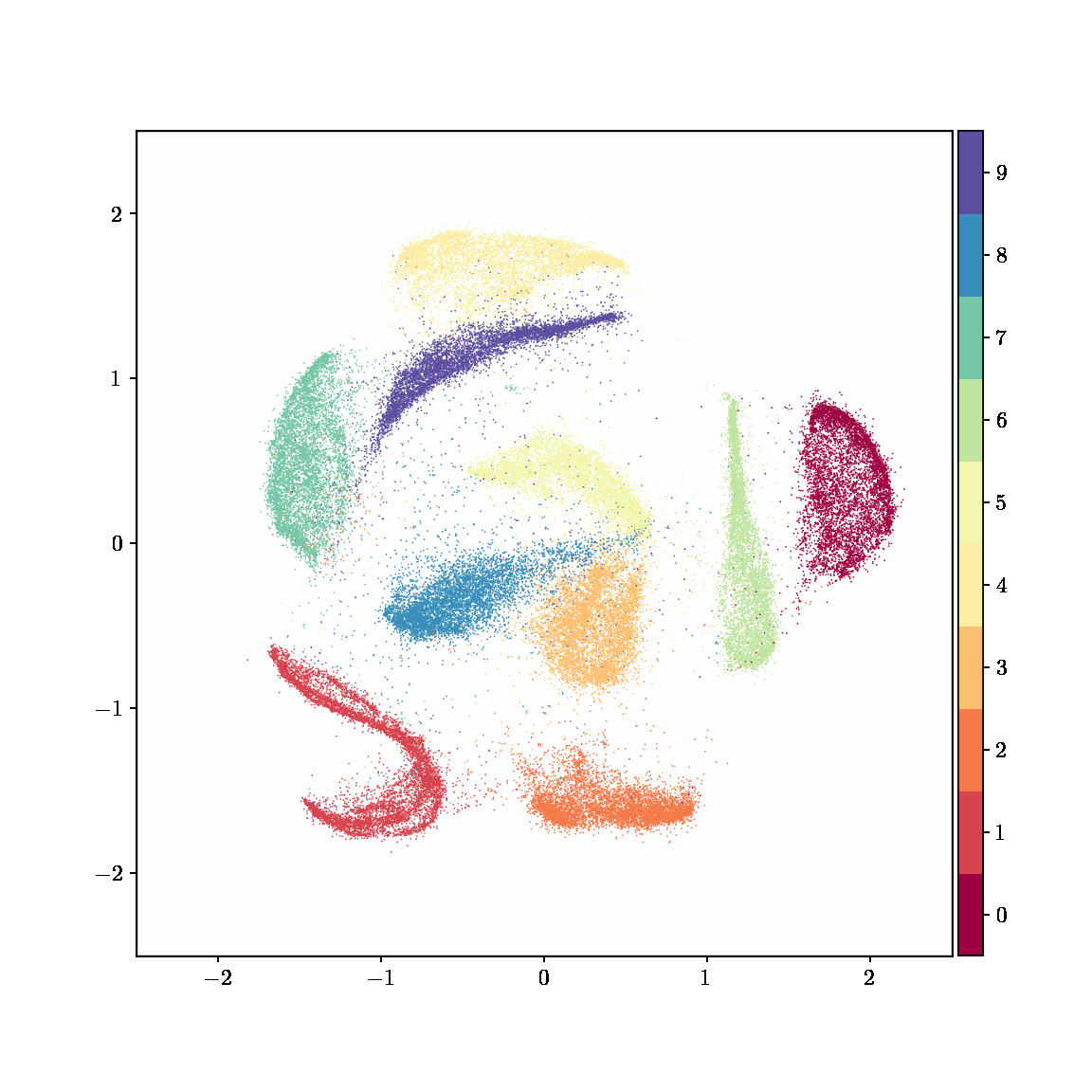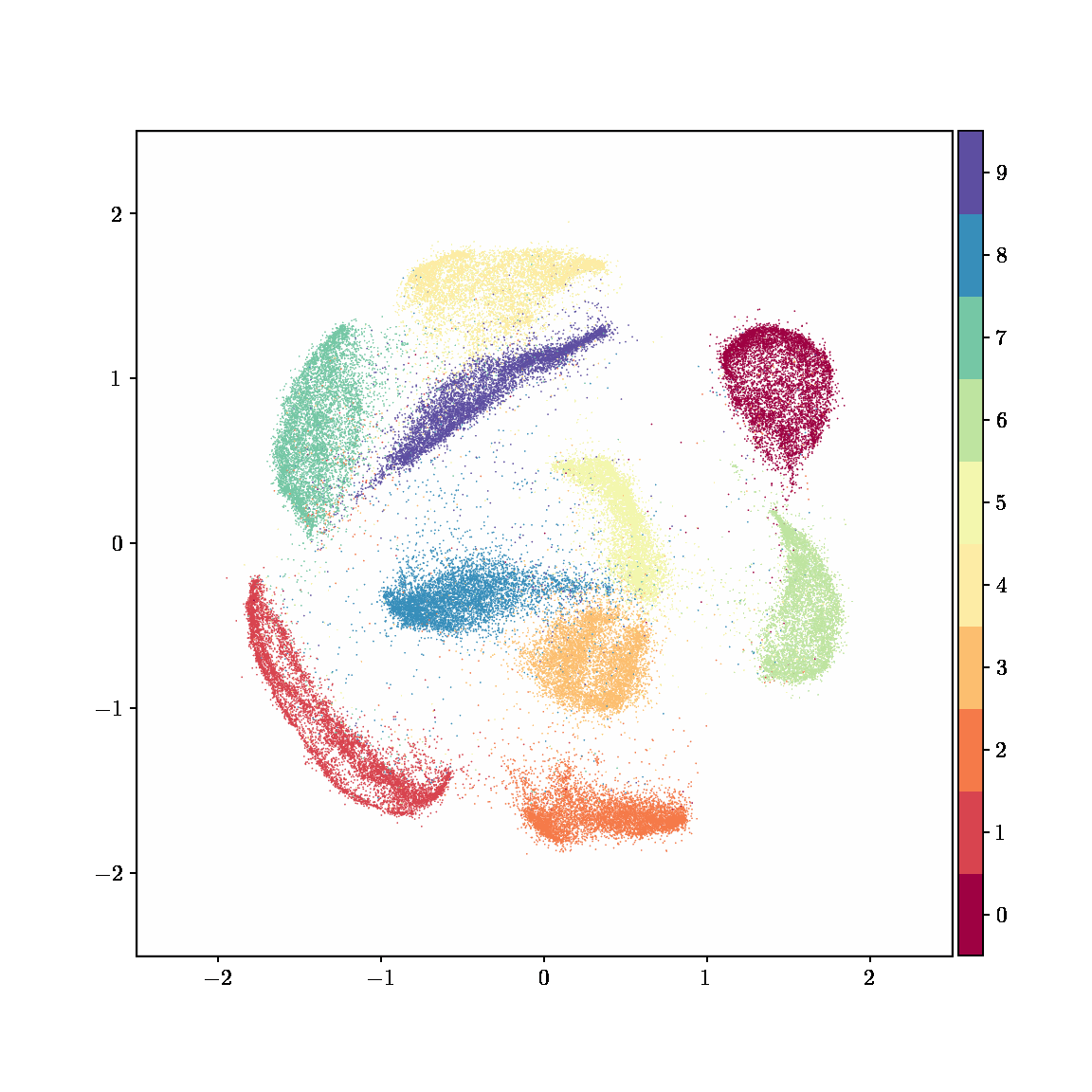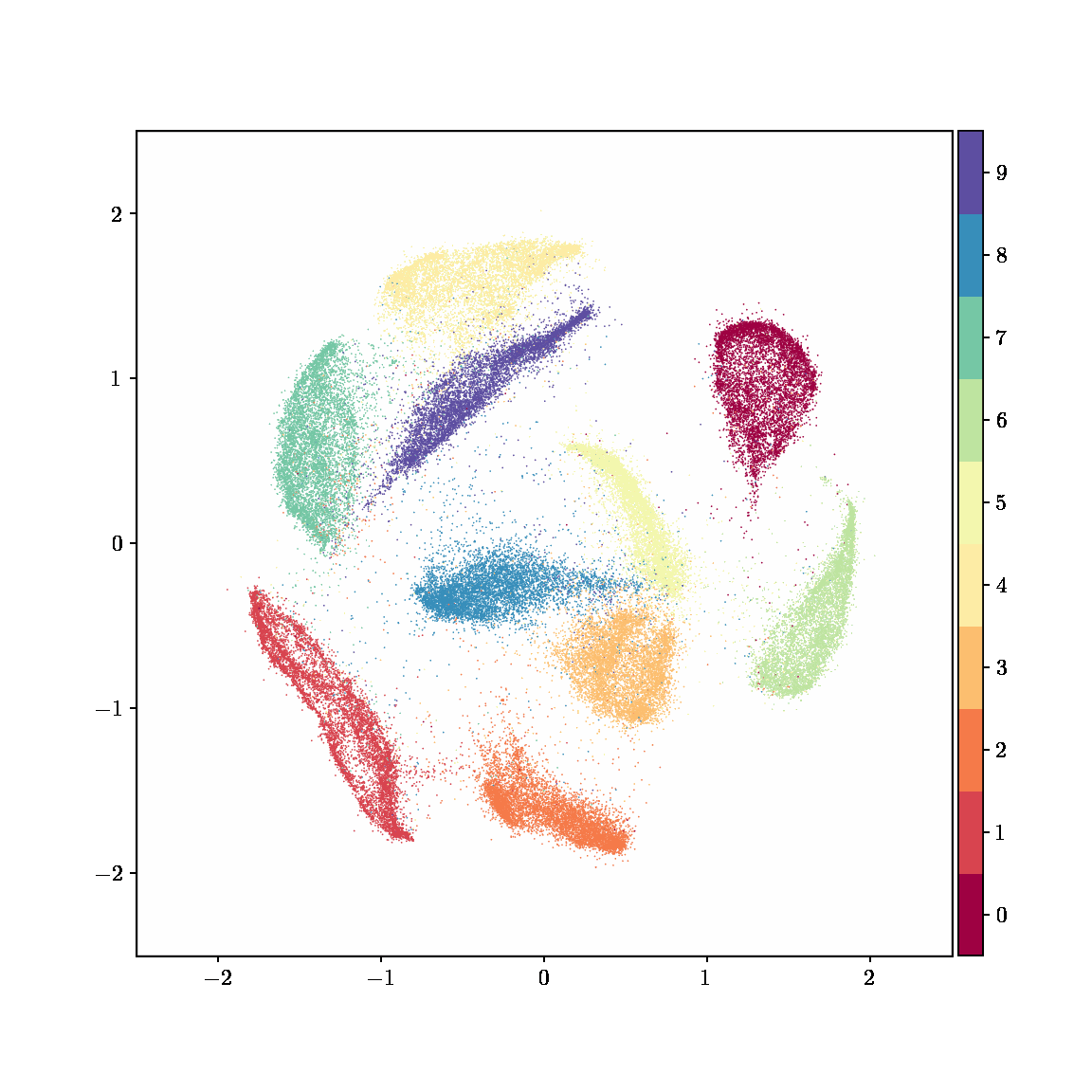
PyMDE is easy to use. It’s easy to get started with PyMDE. With just one line of code, you can embed high-dimensional vectors or the nodes of a graph into any target dimension. Creating custom embeddings is also easy, because PyMDE separates the description of an embedding from how it is computed: you say what kind of embedding you want, and PyMDE will compute it for you.
In addition to computing embeddings, PyMDE can plot them (if the embedding dimension is three or less), create GIFs of the embedding process, help you compare different embeddings, preprocess your data, and more.
PyMDE is principled. The MDE framework provides principled methods to debug and analyze embeddings. These methods can be used to make your embeddings more faithful to your original data, and to search for anomalies and outliers in the original data.
PyMDE is fast, thanks to vecotrization, GPU acceleration, and Rust. PyMDE is competitive in runtime with more specialized embedding methods, like UMAP, and can scale to datasets with millions of items. With a GPU, it can be even faster. PyMDE scales well with the embedding dimension, meaning that you can easily embed into dimensions such as 50, 100, or 250.
PyMDE preprocesses original data matrices and constructs neighborhood graphs and shortest path distances using fast routines implemented in Rust, taking advantage of SIMD, BLAS, and basic parallelism.
PyMDE draws graphs orders of magnitude faster than NetworkX.
PyMDE is easy to install. PyMDE does not depend on Numba, thanks to its Rust-based preprocessing. This means that it does not inherit the limitations that Numba imposes on NumPy and Python versions. Find wheels for all modern Python versions and all architectures on PyPI.